
What is SLCN?
SLCN (Surface Learning for Clinical Neuroimaging) is a new MICCAI workshop aimed at promoting the development of deep learning methods suitable for cortical surface data. This year, participants are tasked with predicting developmental phenotypes (gestational age at birth and cognitive score) from cortical surface data acquired as part of the Developing Human Connectome Project.
This page will be updated regularly to provide information on the submission instructions and evaluation phases. Please, check instructions and email us at slcn.challenge@gmail.com for any question.
The challenge of identifying neurodevelopmental biomarkers
Detecting biomarkers from neuroimaging data is highly challenging, since the degree of topographic variability of cortical shape and functional organisation across individuals limits the accuracy with which brains can be compared through image registration (Glasser et al., 2016; Kong et al., 2019). There is a clear need for precision imaging tools of the brain which are robust to these factors.
Recently, (Fawaz et al., 2021; Dahan et al., 2022) proposed a novel benchmark for prototyping tools for cortical analysis based on the regression of clinical phenotypes from the dHCP (Makropoulos et al., 2018; Bozek et al., 2018) - a unique open dataset of multimodal neonatal brain images, acquired from preterm and term neonates between 24 and 45 weeks’ postmenstrual age. Over this period of late gestation, the human brain undergoes rapid growth and maturation, characterised by highly dynamic and organised cellular and molecular processes (Figure 1). Disruptions to these processes, for example due to preterm birth, can have serious consequences resulting in significant increased risk of those neonates developing neuropsychiatric conditions including autism spectrum disorder, attention deficit hyperactivity disorder (ADHD), schizophrenia, bipolar affective disorder and major depressive disorder in later life (Nosarti et al., 2007; Nosarti et al., 2012).
Tasks
The goal of this challenge will therefore be to elicit submissions of novel methods for cortical phenotype regression, with emphasis on interpretable or explainable machine learning methods which deliver biomarkers predictive of risk for neurodevelopmental impairment. These will be benchmarked on the tasks of:
- Regression of gestational age at birth (seen as a correlate of prematurity).
- Prediction of cognitive outcome measured using Bayley-III composite cognitive scores.
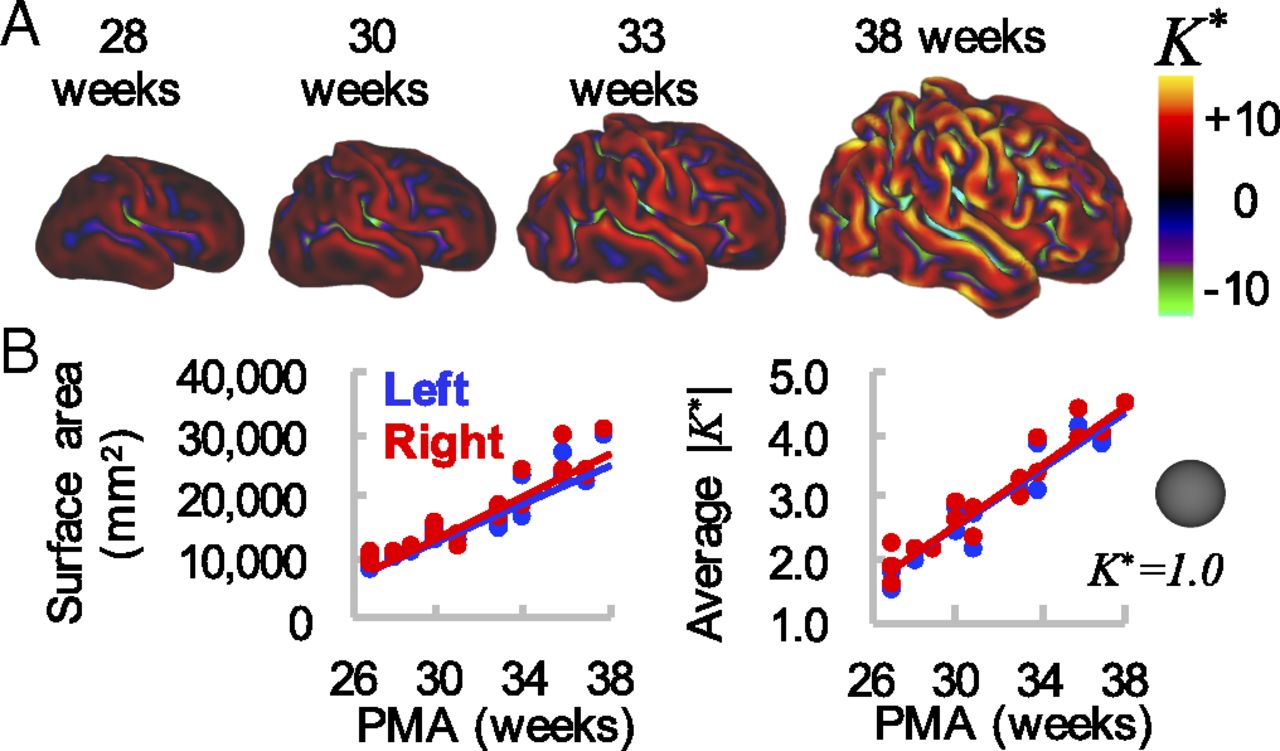 |
|---|
| Figure 1. Global measures of cortical surface area and folding increase over time. (A) Cortical midthickness surfaces for individual hemisphere at 28 wk, 30 wk, 33 wk, and 38 wk PMA. Color overlay represents nondimensional curvature, K, a useful metric of the degree of folding. (B) Total cortical surface area (Left) and average magnitude of K (Right) increase with time. However, these global measures do not provide information about regional variations in maturation and morphology. Adapted from Garcia et al., 2018. |